I’m interrupting the series on cognitive biases (unskilled-and-unaware, superiority illusion, and depressive realism) to tell you that I admit it, I’m old. -Ish. Well, ok, I’m not that old. But this following paper made me feel that old. Because it invalidates some stuff I thought I knew about molecular cell biology. Mind totally blown.
It all started with a paper freshly published two days ago and that I’ll cover tomorrow. It’s about what the title says: mRNA can travel between cells packaged nicely in vesicles and once in a target cell can be made into protein there. I’ll explain – briefly! – why this is such a mind-blowing thing.

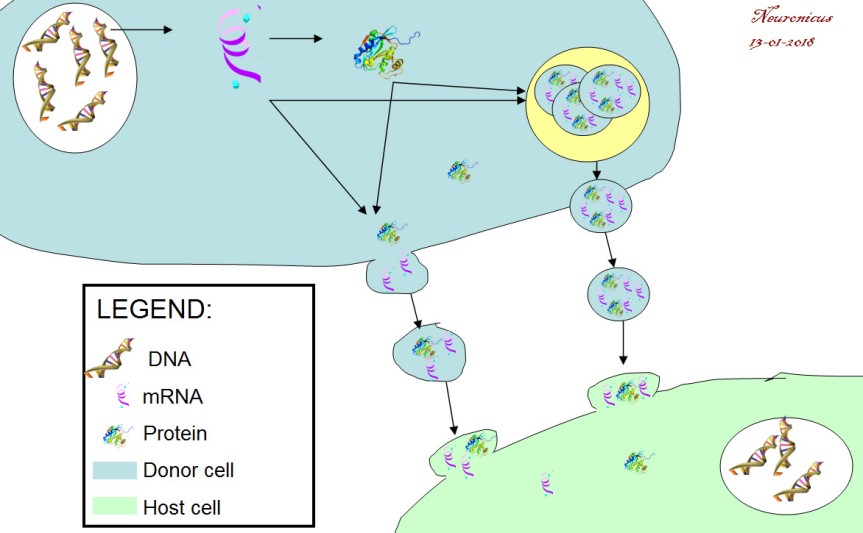
We’ll start with the central dogma of molecular biology (specialists, please bear with me): the DNA is transcribed into RNA and the RNA is translated into protein (see Fig. 1). It is an oversimplification of the complexity of information flow in a biological system, but it’ll do for our purposes.
DNA needs to be transcribed into RNA because RNA is a much more flexible molecule and thus can do many things. So RNA is the traveling mule between DNA and the place where its information becomes protein, i.e. ribosome. Hence the name mRNA. Just kidding; m stands for messenger RNA (not that I will ever be able to call that ever again: muleRNA is stuck in my brain now).
There are many kinds of RNA: some don’t even get out of the nucleus, some are chopped and re-glued (alternative splicing), some decide which bits of DNA (genes) are to be expressed, some are busy housekeepers and so on. Once an RNA has finished its business it is degraded in many inventive ways. It cannot leave the cell because it cannot cross the cell membrane. And that was that. Or so I’ve been taught.
Exceptions from the above were viruses whose ways of going from cell to cell are very clever. A virus is a stretch of nucleic acids (DNA and/or RNA) and some proteins encapsulated in a blob (capsid). Not a cell!
In the ’90s several groups were looking at some blobs (yes, most stuff in biology can be defined by the all-encompassing and enlightening term of ‘blob’) that cells spew out every now and then. These were termed extracellular vesicles (EV) for obvious reasons. Turned out that many kinds of cells were doing it and on a much more regular basis than previously thought. The contents of these EVs varied quite a bit, based on the type of cells studied. Proteins, mostly, and maybe some cytoplasmic debris. In the ’80s it was thought that this was one way for a cell to get rid of trash. But in 1982, Stegmayr & Ronquist showed that prostate cells release some EVs that result in sperm cell motility increase (Raposo & Stoorvogel, 2013) so, clearly, the EVs were more than trash. Soon it became evident that EVs were another way of cell-to-cell communication. (Note to self: the first time intercellular communication by EVs was demonstrated was in 1982, Stegmayr & Ronquist. Maybe I’ll dig out the paper to cover it sometime).
So. In 2005, Baj-Krzyworzeka et al. (2006) looked at some human cancer cells to see what they spew out and for what purpose. They saw that the cancer cells were transferring some of the tumor proteins packaged in EVs to monocytes. For devious purposes, probably. And then they made to what it looks to me like a serious leap in reasoning: since the EVs contain tumor proteins, why wouldn’t they also contain the mRNA for those proteins? My first answer to that would have been: “because it would be rapidly degraded”. And I would have been wrong. To my credit, if the experiment wouldn’t take up too many resources I still would have done it, especially if I would have some random primers lying around the lab. Luckily for the world, I was not in charge with this particular experiment and Baj-Krzyworzeka et al. (2005) proceeded with a real-time PCR (polymerase chain reaction) which showed them that the EVs released by the tumor cells also contained mRNA.
Now the 1 million dollar, stare-in-your-face question was: is this mRNA functional? Meaning, once delivered to the host cell, would it be translated into protein?
Six months later the group answered it. Ratajcza et al. (2006) used embryonic stem cells as the donor cells and hematopoietic progenitor cells as host cells. First, they found out that if you let the donors spit EVs at the hosts, the hosts are faring much better (better survival, upregulated good genes, phosphorylated MAPK to induce proliferation etc.). Next, they looked at the contents of EVs and found out that they contained proteins and mRNA that promote those good things (Wnt-3 protein, mRNA for transcription factors etc.). Next, to make sure that the host cells don’t show this enrichment all of a sudden out of the goodness of their little pluripotent hearts but is instead due to the mRNA from the donor cells, the authors looked at the expression of one of the transcription factors (Oct-4) in the hosts. They used as host a cell line (SKL) that does not express the pluripotent marker Oct-4. So if the hosts express this protein, it must have come only from outside. Lo and behold, they did. This means that the mRNA carried by the EVs is functional (Fig. 2).
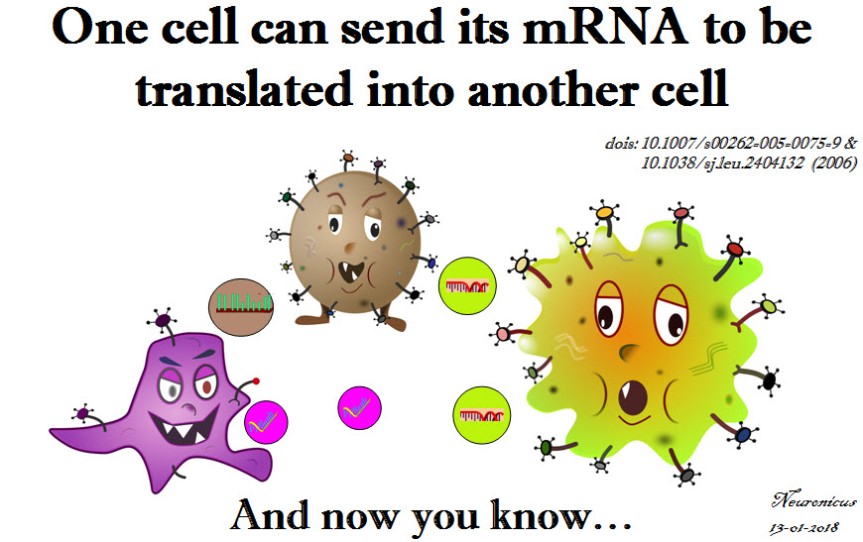
What bugs me is that these papers came out in a period where I was doing some heavy reading. How did I miss this?! Probably because they were published in cancer journals, not my field. But this is big enough you’d think others would mention it. (If you’re a recurrent reader of my blog, by now you should be familiarized with my stream-of-consciousness writing and my admittedly sometimes annoying in-parenthesis-meta-cognitions :D). So how did I miss this? How many more great discoveries have I missed? Am I the only one to discover such fundamental gaps in my knowledge? And thus the imposter syndrome takes root.
Just kidding, I don’t have the imposter syndrome. If anything, I got a superiority illusion complex. And I am absolutely sure that many, many scientists read things they consider fundamental to their way of thinking about the world all the time and wonder what other truly great discoveries are out there already that they missed.
Frankly, I should probably be grateful to this blog – and my friend GT who made me do it – because without nosing outside my field in search of material for it I would have probably remained ignorant of this awesome discovery. So, even if this is a decade old discovery for you, for me is one day old and I am a bit giddy about it.
This is a big deal because of the theoretical implications: a cell’s transcriptome (all the mRNA expressed in a cell) varies not only due to its needs, activity, and experiences, but also due to its neighbors’! A cell is, more or less, its transcriptome. Soooo… if we can change that at will, does that means we can change the type or function of the cell too? There are so many questions that such a discovery raises! And possibilities.
This is also a big deal because it opens up not a new therapy, or a new therapy direction, or a new drug class, but a new DELIVERY METHOD, the Holy Grail of Pharmacopeia. You just put your drug in one of these vesicles and let nature take its course. Of course, there are all sorts of roadblocks to overcome, like specificity, toxicity, etc. Looks like some are already conquered as there are several clinical trials out there that take advantage of this mechanism and I bet there will be more.
Stop by tomorrow for a freshly published paper on this mechanism in neurons.
REFERENCES:
1) Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Urbanowicz B, Brański P, Ratajczak MZ, & Zembala M. (Jul. 2006, Epub 9 Nov 2005). Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunology, Immunotherapy, 55(7):808-818. PMID: 16283305, DOI: 10.1007/s00262-005-0075-9. ARTICLE
2) Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, & Ratajczak MZ (May 2006). Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia, 20(5):847-856. PMID: 16453000, DOI: 10.1038/sj.leu.2404132. ARTICLE | FREE FULLTEXT PDF
Bibliography:
Raposo G & Stoorvogel W. (18 Feb. 2013). Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of Cell Biology, 200(4):373-383. PMID: 23420871, PMCID: PMC3575529, DOI: 10.1083/jcb.201211138. ARTICLE | FREE FULLTEXT PDF
By Neuronicus, 13 January 2018