In the past few days, a new hot subject has gripped the attention of various media and concerned the medical doctors, as if they don’t have enough to deal with: chloroquine. That is because the President of the U.S.A., Donald Trump, endorsed chloroquine as treatment of COVID-19, a “game changer”, despite his very own director of the National Institute of Allergy and Infectious Diseases (NIAID), Dr. Anthony Fauci, very emphatically and vehemently denying that the promise of (hydroxy)chloroquine is beyond anecdotal (see the White House briefing transcript here).
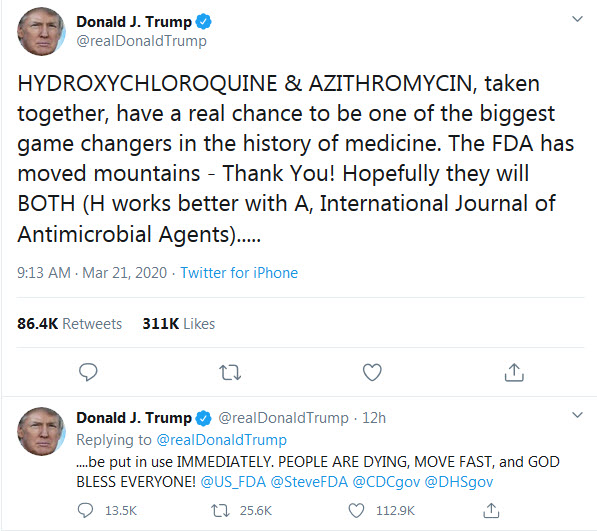
Many medical doctors spoke out urging caution against the drug, particularly against the combination the President endorses: hydroxychloroquine + azithromycin. As I understand it, this combo can be lethal as it can lead to fatal arrhythmia.
As for the (hydroxy)cloroquine’s possibility to help treat COVID-19, the jury is still out. Far out. Meaning that there have been a few interesting observations of the drugs working in a Petri dish (Liu et al. 2020, Wang et al., 2020), but as any pharma company knows, there is a long and perilous way from Petri dishes to pharmacies. To be precise, only 1 in 5000 drugs get from pre-clinical trials to approval and it takes about 12 years for this process to be completed (Kaljevic et al., 2004). The time is so long not because red tape, as some would deplore, but because it takes time to see what it does in humans (Phase 0), what doses are safe and don’t kill you (Phase 1), does it work at all for the intended disease (Phase 2), compare it with other drugs and evaluate the long-term side effects (Phase 3) and, finally, to see the risks and benefits of this drug (Phase 4). While we could probably get rid of Phase 0 and 4 when there is such a pandemic, there is no way I would submit my family to anything that hasn’t passed phases 1, 2, and 3. And those take years. With all the money that a nation-state has, it would still take 18 months to do it semi-properly.
Luckily for all of us, chloroquine is a very old and established anti-malarial medicine, and as such we can safely dispense of Phases 0, 1, and 4, which is fine. So we can start Phase 2 with (hydroxy)chloroquine. And that is exactly what WHO and several others are doing right now. But we don’t have enough data. We haven’t done it yet. So one can hope as much as they want, but that doesn’t make it faster.
Unfortunately – and here we go to the crux of the post -, following the President’s endorsement, many started to hoard chloroquine. Particularly the rich who can afford to “convince” an MD to write them a script for it. In countries where chloroquine is sold without prescription, like Nigeria, where it is used for arthritis, people rushed to clear the pharmacies and some didn’t just stockpiled it, but they took it without reason and without knowing the dosage. And they died. [EDIT, 23 March 2020. If you think that wouldn’t ever happen in the land of the brave, think again, as the first death to irresponsible taking chloroquine just happened in the USA]. In addition, the chloroquine hoarding in US by those who can afford it (is about $200 for 50 pills) lead to lack of supply for those who really need it, like lupus or rheumatology patients.
For those who blindly hoard or take chloroquine without prescription, I have a little morsel of knowledge to impart. Remember I am not an MD; I hold a PhD in neuroscience. So I’ll tell you what my field knows about chloroquine.
Both chloroquine and hydroxychloroquine can cause severe psychosis.
That’s right. More than 7.1 % of people who took chloroquine as prophylaxis or for treatment of malaria developed “mental and neurological manifestations” (Bitta et al., 2017). “Hydroxychloroquine was associated with the highest prevalence of mental neurological manifestations” (p. 12). The phenomenon is well-reported, actually having its own syndrome name: “chloroquine-induced psychosis”. It was observed more than 50 years ago, in 1962 (Mustakallio et al., 1962). The mechanisms are unclear, with several hypotheses being put forward, like the drugs disrupting the NMDA transmission, calcium homeostasis, vacuole exocytosis or some other mysterious immune or transport-related mechanism. Because the symptoms are so acute, so persistent and so diverse than more than one brain neurotransmitter system must be affected.
Chloroquine-induced psychosis has sudden onset, within 1-2 days of ingestion. The syndrome presents with paranoid ideation, persecutory delusions, hallucinations, fear, confusion, delirium, altered mood, personality changes, irritability, insomnia, suicidal ideation, and violence (Biswas et al., 2014, Mascolo et al., 2018). All these at moderately low or therapeutically recommended doses (Good et al., 1982). One or two pills can be lethal in toddlers (Smith & Klein-Schwartz, 2005). The symptoms persist long after the drug ingestion has stopped (Maxwell et al., 2015).
Still want to take it “just in case”?

P.S. A clarification: the chemical difference between hydroxychloroquine and chloroquine is only one hydroxyl group (OH). Both are antimalarial and both have been tested in vitro for COVID-19. There are slight differences between them in terms of toxicity, safety and even mechanisms, but for the intents of this post I have treated them as one drug, since both produce psychosis.
REFERENCES:
1) Biswas PS, Sen D, & Majumdar R. (2014, Epub 28 Nov 2013). Psychosis following chloroquine ingestion: a 10-year comparative study from a malaria-hyperendemic district of India. General Hospital Psychiatry, 36(2): 181–186. doi: 10.1016/j.genhosppsych.2013.07.012, PMID: 24290896 ARTICLE
2) Bitta MA, Kariuki SM, Mwita C, Gwer S, Mwai L, & Newton CRJC (2 Jun 2017). Antimalarial drugs and the prevalence of mental and neurological manifestations: A systematic review and meta-analysis. Version 2. Wellcome Open Research, 2(13): 1-20. PMCID: PMC5473418, PMID: 28630942, doi: 10.12688/wellcomeopenres.10658.2 ARTICLE|FREE FULLTEXT PDF
4) Good MI & Shader RI. Lethality and behavioral side effects of chloroquine (1982). Journal of Clinical Psychopharmacology, 2(1): 40–47. doi: 10.1097/00004714-198202000-00005, PMID: 7040501. ARTICLE
3) Kraljevic S, Stambrook PJ, & Pavelic K (Sep 2004). Accelerating drug discovery. EMBO Reports, 5(9): 837–842. doi: 10.1038/sj.embor.7400236, PMID: 15470377, PMCID: PMC1299137. ARTICLE| FREE FULLTEXT PDF
4) Mascolo A, Berrino PM, Gareri P, Castagna A, Capuano A, Manzo C, & Berrino L. (Oct 2018, Epub 9 Jun 2018). Neuropsychiatric clinical manifestations in elderly patients treated with hydroxychloroquine: a review article. Inflammopharmacology, 26(5): 1141-1149. doi: 10.1007/s10787-018-0498-5, PMID: 29948492. ARTICLE
5) Maxwell NM, Nevin RL, Stahl S, Block J, Shugarts S, Wu AH, Dominy S, Solano-Blanco MA, Kappelman-Culver S, Lee-Messer C, Maldonado J, & Maxwell AJ (Jun 2015, Epub 9 Apr 2015). Prolonged neuropsychiatric effects following management of chloroquine intoxication with psychotropic polypharmacy. Clinical Case Reports, 3(6): 379-87. doi: 10.1002/ccr3.238, PMID: 26185633. ARTICLE | FREE FULLTEXT PDF
6) Mustakallio KK, Putkonen T, & Pihkanen TA (1962 Dec 29). Chloroquine psychosis? Lancet, 2(7270): 1387-1388. doi: 10.1016/s0140-6736(62)91067-x, PMID: 13936884. ARTICLE
7) Smith ER & Klein-Schwartz WJ (May 2005). Are 1-2 dangerous? Chloroquine and hydroxychloroquine exposure in toddlers. The Journal of Emergency Medicine, 28(4): 437-443. doi: 10.1016/j.jemermed.2004.12.011, PMID: 15837026. ARTICLE
Studies about chloroquine and hydoxychloroquine on SARS-Cov2 in vitro:
- Liu, J., Cao, R., Xu, M., Wang, X., Zhang, H., Li, Y., Hu, Z., Zhong, W., & Wang, M. (18 March 2020). Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discovery, 6 (16), https://doi.org/10.1038/s41421-020-0156-0 ARTICLE | FREE FULLTEXT PDF
- Wang, M., Cao, R., Zhang, H., Yang, X., Liu, J., Xu, M., Shi, Z., Hu, Z., Zhong, W., & Xiao, G. (18 March 2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30: 269–271. https://doi.org/10.1038/s41422-020-0282-0. ARTICLE | FREE FULLTEXT PDF
First study about chloroquine and hydoxychloroquine on SARS-Cov2 in vivo below. Unfortunately, it has some methodological flaws, see here and here, which hopefully will be corrected once the peer-reviewers will take a closer look at it. UPDATE [31-3-2020]: It seems the article is flawed in more than one way, with serious ethical issues (timeline of treatment doesn’t match the methods reported, patients appear and disappear from data points, graphs different depending on the venue, published in the same journal where the author is editor, no peer-review, no blind, no placebo, controls barely tested, plus, no reputable researcher should announce that they are the genius that cured COVID-19 on a YouTube video, particularly when they get to publish in 24 hours anyway).
Anyway, here it is:
- Gautret P, Lagier J-C, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Esteves Vieira V, Tissot Dupont H,Colson SEP, Chabriere E, La Scola B, Rolain J-M, Brouqui P, Raoult D. (20 March 2020). Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. International Journal of Antimicrobial Agents, PII:S0924-8579(20)30099-6, https://doi.org/10.1016/j.ijantimicag.2020.105949. ARTICLE | FREE FULLTEXT PDF
These studies are also not peer reviewed or at the very least not properly peer reviewed. I say that so as to take them with a grain of salt. Not to criticize in the slightest. Because I do commend the speed with which these were done and published given the pandemic. Bravo to all the authors involved (except maybe the last one f it proves to be fraudulent). And also a thumbs up to the journals which made the data freely available in record time. Unfortunately, from these papers to a treatment we still have a long way to go.
By Neuronicus, 22 March 2020


 Excerpt from Walker et al. (2017), p. 5:
Excerpt from Walker et al. (2017), p. 5:










 The title of today’s post wouldn’t make any sense for anybody who isn’t a preschooler’s parent or teacher in the USA. You see, on the west side of the Atlantic there is a debate on whether a play-based curriculum for a preschool is more advantageous than a more academic-based one. Preschool age is 3 to 4 years; kindergarten starts at 5.
The title of today’s post wouldn’t make any sense for anybody who isn’t a preschooler’s parent or teacher in the USA. You see, on the west side of the Atlantic there is a debate on whether a play-based curriculum for a preschool is more advantageous than a more academic-based one. Preschool age is 3 to 4 years; kindergarten starts at 5.







 When you’re the only scientist in the family you get asked the weirdest things. Actually, I’m not the only one, but the other one is a chemist and he’s mostly asked about astrophysics stuff, so he doesn’t really count, because I am the one who gets asked about rare diseases and medication side-effects and food advice. Never mind that I am a neuroscientist and I have professed repeatedly and quite loudly my minimum knowledge of everything from the neck down, all eyes turn to me when the new arthritis medication or the unexpected side-effects of that heart drug are being brought up. But, curiously, if I dare speak about brain stuff I get the looks that a thing the cat just dragged in gets. I guess everybody is an expert on how the brain works on account of having and using one, apparently. Everybody, but the actual neuroscience expert whose input on brain and behavior is to be tolerated and taken with a grain of salt at best, but whose opinion on stomach distress is of the utmost importance and must be listened to reverentially in utter silence [eyes roll].
When you’re the only scientist in the family you get asked the weirdest things. Actually, I’m not the only one, but the other one is a chemist and he’s mostly asked about astrophysics stuff, so he doesn’t really count, because I am the one who gets asked about rare diseases and medication side-effects and food advice. Never mind that I am a neuroscientist and I have professed repeatedly and quite loudly my minimum knowledge of everything from the neck down, all eyes turn to me when the new arthritis medication or the unexpected side-effects of that heart drug are being brought up. But, curiously, if I dare speak about brain stuff I get the looks that a thing the cat just dragged in gets. I guess everybody is an expert on how the brain works on account of having and using one, apparently. Everybody, but the actual neuroscience expert whose input on brain and behavior is to be tolerated and taken with a grain of salt at best, but whose opinion on stomach distress is of the utmost importance and must be listened to reverentially in utter silence [eyes roll].




