A few days ago, a follower of mine gave me an interesting read from The Atlantic regarding the dinosaur extinction. Like many of my generation, I was taught in school that dinosaurs died because an asteroid hit the Earth. That led to a nuclear winter (or a few years of ‘nuclear winters’) which killed the photosynthetic organisms, and then the herbivores didn’t have anything to eat so they died and then the carnivores didn’t have anything to eat and so they died. Or, as my 4-year-old puts it, “[in a solemn voice] after the asteroid hit, big dusty clouds blocked the sun; [in an ominous voice] each day was colder than the previous one and so, without sunlight to keep them alive [sad face, head cocked sideways], the poor dinosaurs could no longer survive [hands spread sideways, hung head] “. Yes, I am a proud parent. Now I have to do a sit-down with the child and explain that… What, exactly?
Well, The Atlantic article showcases the struggles of a scientist – paleontologist and geologist Gerta Keller – who doesn’t believe the mainstream asteroid hypothesis; rather she thinks there is enough evidence to point out that extreme volcano eruptions, like really extreme, thousands of times more powerful than anything we know in the recorded history, put out so much poison (soot, dust, hydrofluoric acid, sulfur, carbon dioxide, mercury, lead, and so on) in the atmosphere that, combined with the consequent dramatic climate change, killed the dinosaurs. The volcanoes were located in India and they erupted for hundreds of thousands of years, but most violent eruptions, Keller thinks, were in the last 40,000 years before the extinction. This hypothesis is called the Deccan volcanism from the region in India where these nasty volcanoes are located, first proposed by Vogt (1972) and Courtillot et al. (1986).

So which is true? Or, rather, because this is science we’re talking about, which hypothesis is more supported by the facts: the volcanism or the impact?
The impact hypothesis was put forward in 1980 when Walter Alvarez, a geologist, noticed a thin layer of clay in rocks that were about 65 million years old, which coincided with the time when the dinosaurs disappeared. This layer is on the KT boundary (sometimes called K-T, K-Pg, or KPB, looks like the biologists are not the only ones with acronym problems) and marks the boundary between the Cretaceous and Paleogenic geological periods (T is for Triassic, yeah, I know). Walter asked his father, the famous Nobel Prize physicist Louis Alvarez, to take a look at it and see what it is. Alvarez Sr. analyzed it and decided that the clay contains a lot of iridium, dozens of times more than expected. After gathering more samples from Europe and New Zealand, they published a paper (Alvarez et al., 1980) in which the scientists reasoned that because Earth’s iridium is deeply buried in its bowels and not in its crust, this iridium at the K-Pg boundary is of extraterrestrial origin, which could be brought here only by an asteroid/comet. This is also the paper in which it was put forth for the first time the conjecture that the asteroid impact killed the dinosaurs, based on the uncanny coincidence of timing.

The discovery of the Chicxulub crater in Mexico followed a more sinuous path because the geophysicists who first discovered it in the ’70s were working for an oil company, looking for places to drill. Once the dinosaur-died-due-to-asteroid-impact hypothesis gained popularity outside academia, the geologists and the physicists put two-and-two together, acquired more data, and published a paper (Hildebrand et al., 1991) where the Chicxulub crater was for the first time linked with the dinosaur extinction. Although the crater was not radiologically dated yet, they had enough geophysical, stratigraphic, and petrologic evidence to believe it was as old as the iridium layer and the dinosaur die-out.
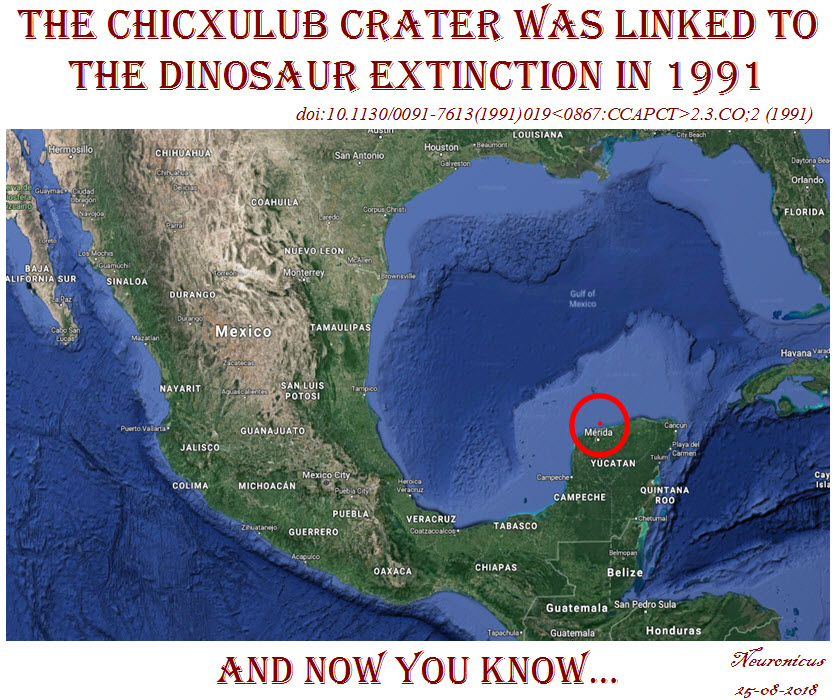
But, devil is in the details, as they say. Keller published a paper in 2007 saying the Chicxulub event predates the extinction by some 300,000 years (Keller et al., 2007). She looked at geological samples from Texas and found the glass granule layer (indicator of the Chicxhulub impact) way below the K-Pg boundary. So what’s up with the iridium then? Keller (2014) believes that is not of extraterrestrial origin and it might well have been spewed up by a particularly nasty eruption or the sediments got shifted. Schulte et al. (2010), on the other hand, found high levels of iridium in 85 samples from all over the world in the KPG layer. Keller says that some other 260 samples don’t have iridium anomalies. As a response, Esmeray-Senlet et al. (2017) used some fancy Mass Spectrometry to show that the iridium profiles could have come only from Chicxulub, at least in North America. They argue that the variability in iridium profiles around the world is due to regional geochemical processes. And so on, and so on, the controversy continues.
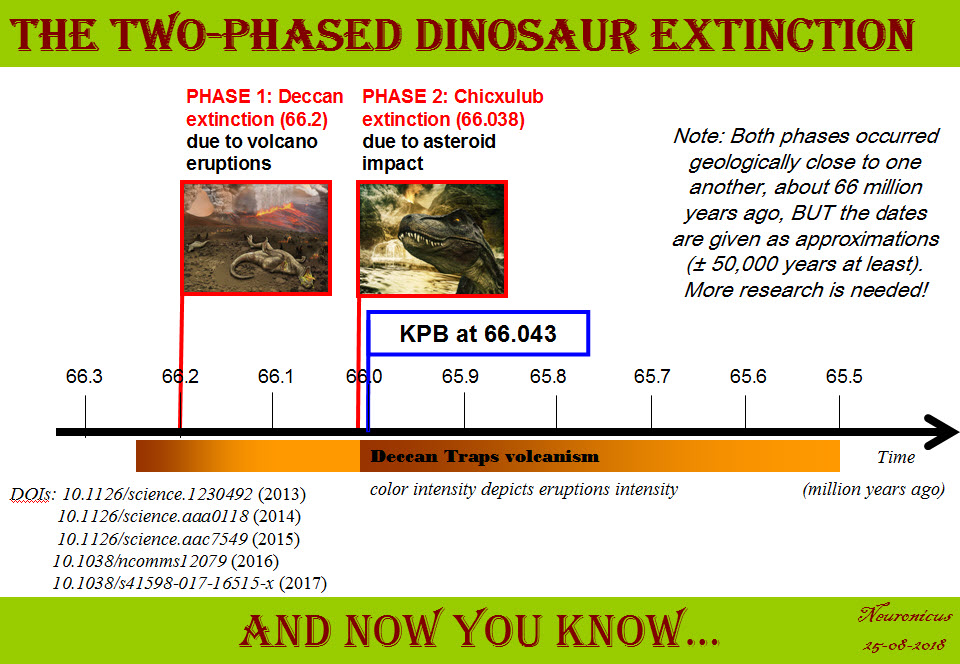
Actual radioisotope dating was done a bit later in 2013: date of K-Pg is 66.043 ± 0.043 MA (millions of years ago), date of the Chicxulub crater is 66.038 ±.025/0.049 MA. Which means that the researchers “established synchrony between the Cretaceous-Paleogene boundary and associated mass extinctions with the Chicxulub bolide impact to within 32,000 years” (Renne et al., 2013), which is a blink of an eye in geological times.

Now I want you to understand that often in science, though by far not always, matters are not so simple as she is wrong, he is right. In geology, what matters most is the sample. If the sample is corrupted, so will be your conclusions. Maybe Keller’s or Renne’s samples were affected by a myriad possible variables, some as simple as shifting the dirt from here to there by who knows what event. After all, it’s been 66 million years since. Also, methods used are just as important and dating something that happened so long ago is extremely difficult due to intrinsic physical methodological limitations. Keller (2014), for example, claims that Renne couldn’t have possibly gotten such an exact estimation because he used Argon isotopes when only U-Pb isotope dilution–thermal ionization mass spectrometry (ID-TIMS) zircon geochronology could be so accurate. But yet again, it looks like he did use both, so… I dunno. As the over-used always-trite but nevertheless extremely important saying goes: more data is needed.
Even if the dating puts Chicxulub at the KPB, the volcanologists say that the asteroid, by itself, couldn’t have produced a mass extinction because there are other impacts of its size and they did not have such dire effects, but were barely noticeable at the biota scale. Besides, most of the other mass extinctions on the planet have been already associated with extreme volcanism (Archibald et al., 2010). On the other hand, the circumstances of this particular asteroid could have made it deadly: it landed in the hydrocarbon-rich areas that occupied only 13% of the Earth’s surface at the time which resulted in a lot of “stratospheric soot and sulfate aerosols and causing extreme global cooling and drought” (Kaiho & Oshima, 2017). Food for thought: this means that the chances of us, humans, to be here today are 13%!…
I hope that you do notice that these are very recent papers, so the issue is hotly debated as we speak.
It is possible, nay probable, that the Deccan volcanism, which was going on long before and after the extinction, was exacerbated by the impact. This is exactly what Renne’s team postulated in 2015 after dating the lava plains in the Deccan Traps: the eruptions intensified about 50,000 years before the KT boundary, from “high-frequency, low-volume eruptions to low-frequency, high-volume eruptions”, which is about when the asteroid hit. Also, the Deccan eruptions continued for about half a million years after KPB, “which is comparable with the time lag between the KPB and the initial stage of ecological recovery in marine ecosystems” (Renne et al., 2016, p. 78).
Since we cannot get much more accurate dating than we already have, perhaps the fossils can tell us whether the dinosaurs died abruptly or slowly. Because if they got extinct in a few years instead of over 50,000 years, that would point to a cataclysmic event. Yes, but which one, big asteroid or violent volcano? Aaaand, we’re back to square one.
Actually, the last papers on the matter points to two extinctions: the Deccan extinction and the Chicxulub extinction. Petersen et al., (2016) went all the way to Antarctica to find pristine samples. They noticed a sharp increase in global temperatures by about 7.8 ºC at the onset of Deccan volcanism. This climate change would surely lead to some extinctions, and this is exactly what they found: out of 24 species of marine animals investigated, 10 died-out at the onset of Deccan volcanism and the remaining 14 died-out when Chicxulub hit.
In conclusion, because this post is already verrrry long and is becoming a proper college review, to me, a not-a-geologist/paleontologist/physicist-but-still-a-scientist, things happened thusly: first Deccan traps erupted and that lead to a dramatic global warming coupled with spewing poison in the atmosphere. Which resulted in a massive die-out (about 200,000 years before the bolide impact, says a corroborating paper, Tobin, 2017). The surviving species (maybe half or more of the biota?) continued the best they could for the next few hundred thousand years in the hostile environment. Then the Chicxulub meteorite hit and the resulting megatsunami, the cloud of super-heated dust and soot, colossal wildfires and earthquakes, acid rain and climate cooling, not to mention the intensification of the Deccan traps eruptions, finished off the surviving species. It took Earth 300,000 to 500,000 years to recover its ecosystem. “This sequence of events may have combined into a ‘one-two punch’ that produced one of the largest mass extinctions in Earth history” (Petersen et al., 2016, p. 6).
By Neuronicus, 25 August 2018
P. S. You, high school and college students who will use this for some class assignment or other, give credit thusly: Neuronicus (Aug. 26, 2018). The FIRSTS: The cause(s) of dinosaur extinction. Retrieved from https://scientiaportal.wordpress.com/2018/08/26/the-firsts-the-causes-of-dinosaur-extinction/ on [date]. AND READ THE ORIGINAL PAPERS. Ask me for .pdfs if you don’t have access, although with sci-hub and all… not that I endorse any illegal and fraudulent use of the above mentioned server for the purpose of self-education and enlightenment in the quest for knowledge that all academics and scientists praise everywhere around the Globe!
EDIT March 29, 2019. Astounding one-of-a-kind discovery is being brought to print soon. It’s about a site in North Dakota that, reportedly, has preserved the day of the Chicxhulub impact in amazing detail, with tons of fossils of all kinds (flora, mammals, dinosaurs, fish) which seems to put the entire extinction of dinosaurs in one day, thus favoring the asteroid impact hypothesis. The data is not out yet. Can’t wait til it is! Actually, I’ll have to wait some more after it’s out for the experts to examine it and then I’ll find out. Until then, check the story of the discovery here and here.
REFERENCES:
1. Alvarez LW, Alvarez W, Asaro F, & Michel HV (6 Jun 1980). Extraterrestrial cause for the cretaceous-tertiary extinction. PMID: 17783054. DOI: 10.1126/science.208.4448.1095 Science, 208(4448):1095-1108. ABSTRACT | FULLTEXT PDF
2. Archibald JD, Clemens WA, Padian K, Rowe T, Macleod N, Barrett PM, Gale A, Holroyd P, Sues HD, Arens NC, Horner JR, Wilson GP, Goodwin MB, Brochu CA, Lofgren DL, Hurlbert SH, Hartman JH, Eberth DA, Wignall PB, Currie PJ, Weil A, Prasad GV, Dingus L, Courtillot V, Milner A, Milner A, Bajpai S, Ward DJ, Sahni A. (21 May 2010) Cretaceous extinctions: multiple causes. Science,328(5981):973; author reply 975-6. PMID: 20489004, DOI: 10.1126/science.328.5981.973-aScience. FULL REPLY
3. Courtillot V, Besse J, Vandamme D, Montigny R, Jaeger J-J, & Cappetta H (1986). Deccan flood basalts at the Cretaceous/Tertiary boundary? Earth and Planetary Science Letters, 80(3-4), 361–374. doi: 10.1016/0012-821x(86)90118-4. ABSTRACT
4. Esmeray-Senlet, S., Miller, K. G., Sherrell, R. M., Senlet, T., Vellekoop, J., & Brinkhuis, H. (2017). Iridium profiles and delivery across the Cretaceous/Paleogene boundary. Earth and Planetary Science Letters, 457, 117–126. doi:10.1016/j.epsl.2016.10.010. ABSTRACT
5. Hildebrand AR, Penfield GT, Kring DA, Pilkington M, Camargo AZ, Jacobsen SB, & Boynton WV (1 Sept. 1991). Chicxulub Crater: A possible Cretaceous/Tertiary boundary impact crater on the Yucatán Peninsula, Mexico. Geology, 19 (9): 867-871. DOI: https://doi.org/10.1130/0091-7613(1991)019<0867:CCAPCT>2.3.CO;2. ABSTRACT
6. Kaiho K & Oshima N (9 Nov 2017). Site of asteroid impact changed the history of life on Earth: the low probability of mass extinction. Scientific Reports,7(1):14855. PMID: 29123110, PMCID: PMC5680197, DOI:10.1038/s41598-017-14199-x. . ARTICLE | FREE FULLTEXT PDF
7. Keller G, Adatte T, Berner Z, Harting M, Baum G, Prauss M, Tantawy A, Stueben D (30 Mar 2007). Chicxulub impact predates K–T boundary: New evidence from Brazos, Texas, Earth and Planetary Science Letters, 255(3–4): 339-356. DOI: 10.1016/j.epsl.2006.12.026. ABSTRACT
8. Keller, G. (2014). Deccan volcanism, the Chicxulub impact, and the end-Cretaceous mass extinction: Coincidence? Cause and effect? Geological Society of America Special Papers, 505:57–89. doi:10.1130/2014.2505(03) ABSTRACT
9. Petersen SV, Dutton A, & Lohmann KC. (5 Jul 2016). End-Cretaceous extinction in Antarctica linked to both Deccan volcanism and meteorite impact via climate change. Nature Communications, 7:12079. doi: 10.1038/ncomms12079. PMID: 27377632, PMCID: PMC4935969, DOI: 10.1038/ncomms12079. ARTICLE | FREE FULLTEXT PDF
10. Renne PR, Deino AL, Hilgen FJ, Kuiper KF, Mark DF, Mitchell WS 3rd, Morgan LE, Mundil R, & Smit J (8 Feb 2013). Time scales of critical events around the Cretaceous-Paleogene boundary. Science, 8;339(6120):684-687. doi: 10.1126/science.1230492. PMID: 23393261, DOI: 10.1126/science.1230492 ABSTRACT
11. Renne PR, Sprain CJ, Richards MA, Self S, Vanderkluysen L, Pande K. (2 Oct 2015). State shift in Deccan volcanism at the Cretaceous-Paleogene boundary, possibly induced by impact. Science, 350(6256):76-8. PMID: 26430116. DOI: 10.1126/science.aac7549 ABSTRACT
12. Schoene B, Samperton KM, Eddy MP, Keller G, Adatte T, Bowring SA, Khadri SFR, & Gertsch B (2014). U-Pb geochronology of the Deccan Traps and relation to the end-Cretaceous mass extinction. Science, 347(6218), 182–184. doi:10.1126/science.aaa0118. ARTICLE
13. Schulte P, Alegret L, Arenillas I, Arz JA, Barton PJ, Bown PR, Bralower TJ, Christeson GL, Claeys P, Cockell CS, Collins GS, Deutsch A, Goldin TJ, Goto K, Grajales-Nishimura JM, Grieve RA, Gulick SP, Johnson KR, Kiessling W, Koeberl C, Kring DA, MacLeod KG, Matsui T, Melosh J, Montanari A, Morgan JV, Neal CR, Nichols DJ, Norris RD, Pierazzo E,Ravizza G, Rebolledo-Vieyra M, Reimold WU, Robin E, Salge T, Speijer RP, Sweet AR, Urrutia-Fucugauchi J, Vajda V, Whalen MT, Willumsen PS.(5 Mar 2010). The Chicxulub asteroid impact and mass extinction at the Cretaceous-Paleogene boundary. Science, 327(5970):1214-8. PMID: 20203042, DOI: 10.1126/science.1177265. ABSTRACT
14. Tobin TS (24 Nov 2017). Recognition of a likely two phased extinction at the K-Pg boundary in Antarctica. Scientific Reports, 7(1):16317. PMID: 29176556, PMCID: PMC5701184, DOI: 10.1038/s41598-017-16515-x. ARTICLE | FREE FULLTEXT PDF
15. Vogt, PR (8 Dec 1972). Evidence for Global Synchronism in Mantle Plume Convection and Possible Significance for Geology. Nature, 240(5380), 338–342. doi:10.1038/240338a0 ABSTRACT